ibis: mutate and more aggregates¶
Learning Goals¶
use
group_by()andaggregate()patterns to summarize datause
mutate()to create an new column that is a function of data from existing columns in a table.
import ibis
from ibis import _
import ibis.selectors as s
import seaborn.objects as so
con = ibis.duckdb.connect()
base_url = "https://huggingface.co/datasets/cboettig/ram_fisheries/resolve/main/v4.65/"
stock = con.read_csv(base_url + "stock.csv", nullstr="NA")
timeseries = con.read_csv(base_url + "timeseries.csv", nullstr="NA")
Last time we reached the conclusion that we wanted to average across multiple assessments:
cod_stocks = (
timeseries
.join(stock, "stockid")
.filter(_.tsid == "TCbest-MT")
.filter(_.commonname == "Atlantic cod")
.group_by(_.tsyear, _.stockid, _.primary_country, _.primary_FAOarea)
.agg(catch = _.tsvalue.mean())
)
cod_stocks
r0 := DatabaseTable: ibis_read_csv_oqvhqgdgtvffpdkwj2e2hhgkmi
assessid string
stockid string
stocklong string
tsid string
tsyear int64
tsvalue float64
r1 := DatabaseTable: ibis_read_csv_jvtzdli36jbcdkhuwkurh7rv7y
stockid string
tsn int64
scientificname string
commonname string
areaid string
stocklong string
region string
primary_country string
primary_FAOarea int64
ISO3_code string
GRSF_uuid string
GRSF_areaid string
inmyersdb int64
myersstockid string
state string
r2 := JoinChain[r0]
JoinLink[inner, r1]
r0.stockid == r1.stockid
values:
assessid: r0.assessid
stockid: r0.stockid
stocklong: r0.stocklong
tsid: r0.tsid
tsyear: r0.tsyear
tsvalue: r0.tsvalue
tsn: r1.tsn
scientificname: r1.scientificname
commonname: r1.commonname
areaid: r1.areaid
stocklong_right: r1.stocklong
region: r1.region
primary_country: r1.primary_country
primary_FAOarea: r1.primary_FAOarea
ISO3_code: r1.ISO3_code
GRSF_uuid: r1.GRSF_uuid
GRSF_areaid: r1.GRSF_areaid
inmyersdb: r1.inmyersdb
myersstockid: r1.myersstockid
state: r1.state
r3 := Filter[r2]
r2.tsid == 'TCbest-MT'
r4 := Filter[r3]
r3.commonname == 'Atlantic cod'
Aggregate[r4]
groups:
tsyear: r4.tsyear
stockid: r4.stockid
primary_country: r4.primary_country
metrics:
catch: Mean(r4.tsvalue)
This is great, but we have a lot of individual cod stocks:
(
so.Plot(cod_stocks,
x = "tsyear",
y="catch",
color = "stockid")
.add(so.Lines(linewidth=3))
.layout(size=(12, 8))
)
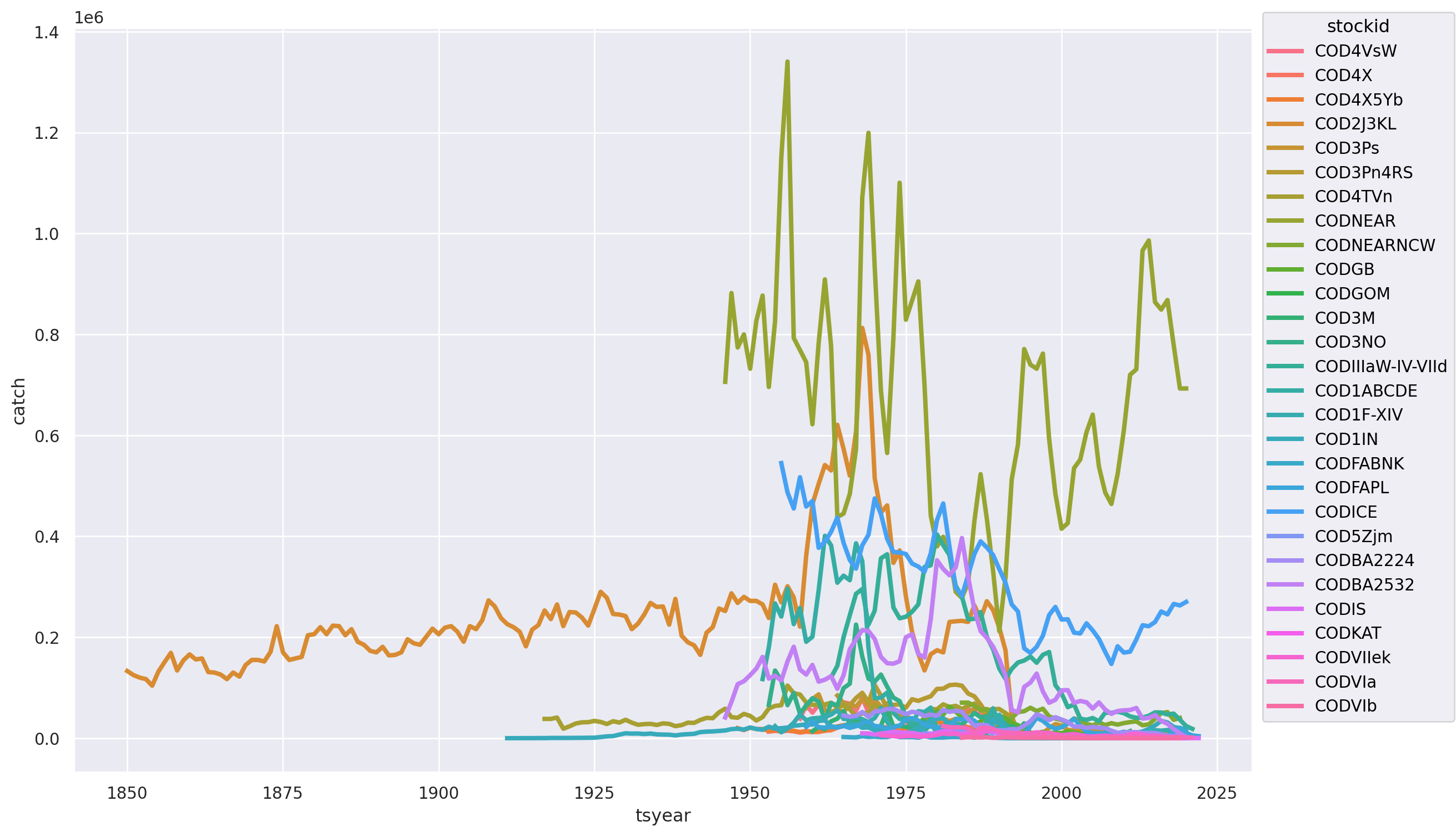
Our good friend, the COD2J3KL series shows up with it’s remarkable declines, but what’s going on with those highly variable but very large catches? This will obviously impact our assessment of whether or not the species as a whole has collapsed. This is too many stocks to easily explore, let’s try breakint this out by at the by country:
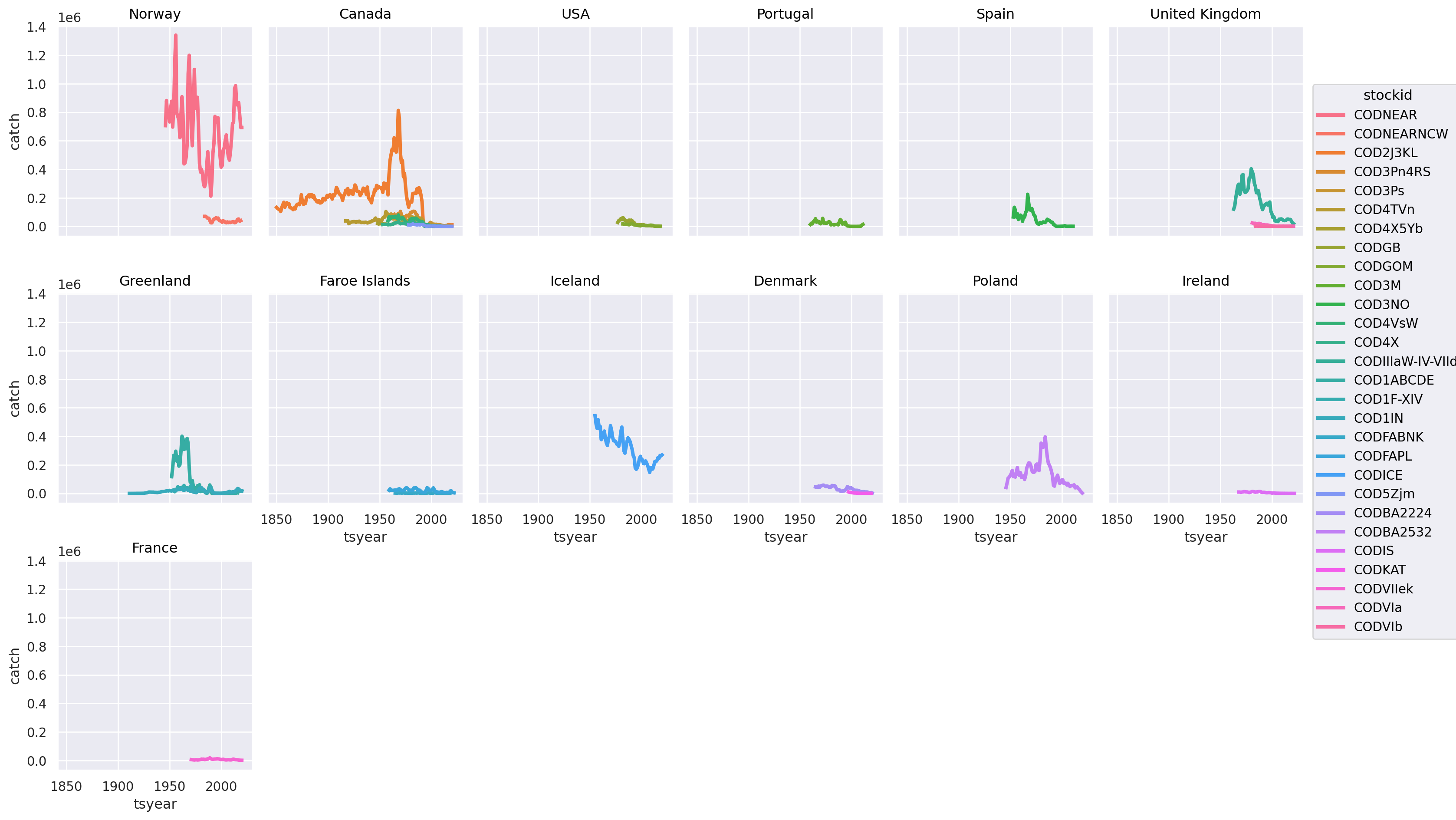
(
so.Plot(cod_stocks,
x = "tsyear",
y="catch",
color = "stockid",
group = "primary_country")
.add(so.Lines(linewidth=3))
.facet("primary_country", wrap = 6)
.layout(size=(16, 10))
)
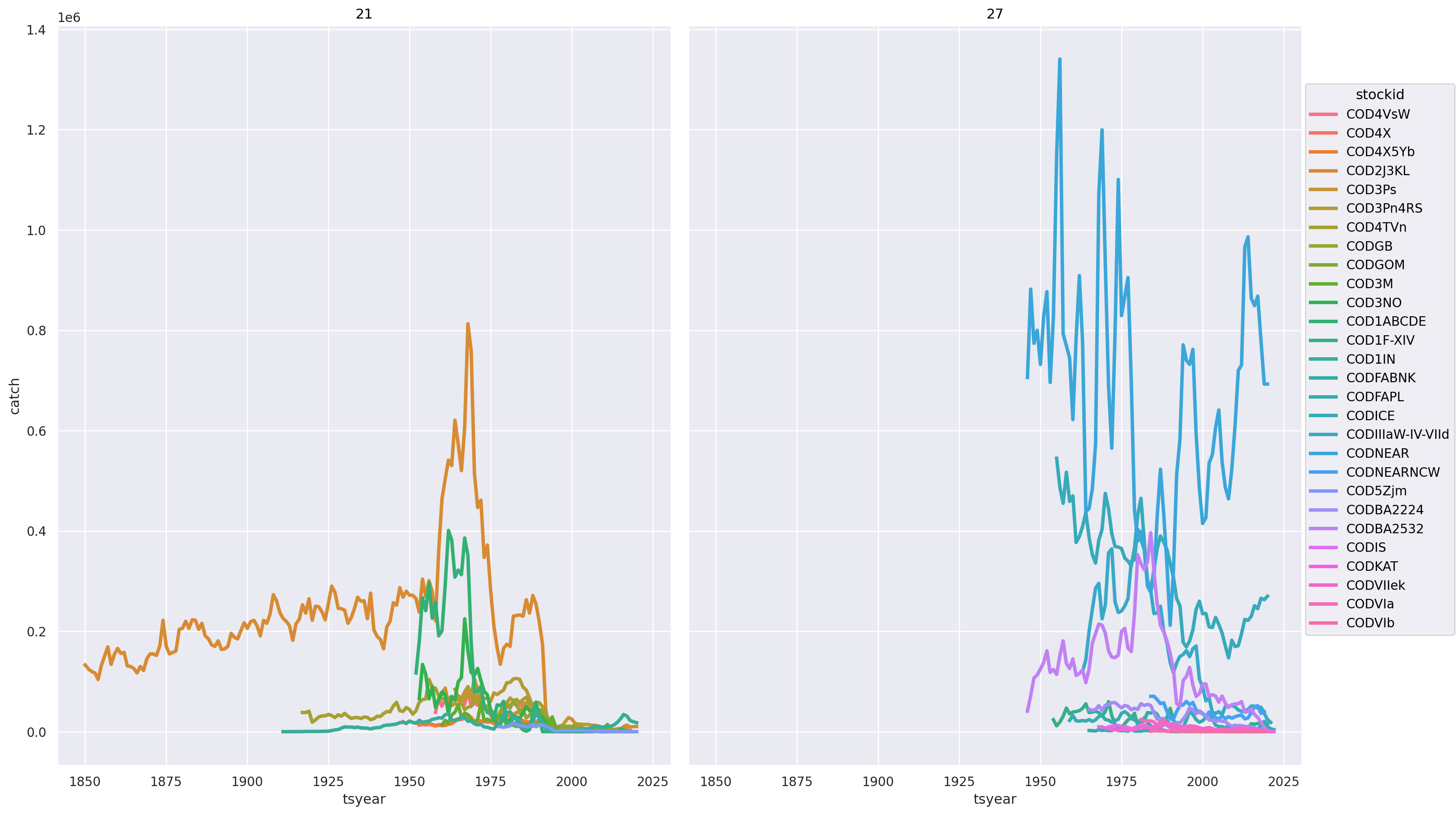
(
so.Plot(cod_stocks,
x = "tsyear",
y="catch",
color = "stockid",
group = "primary_FAOarea")
.add(so.Lines(linewidth=3))
.facet("primary_FAOarea", wrap = 6)
.layout(size=(16, 10))
)
cod_stocks